
Publications
Hu, Y., Morichaud, Z., Perumal, A.S., Roquet-Baneres, F., Brodolin, K., 2014. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic acids research 42, 10399-10408 MoreLess

RbpA, a transcriptional activator that is essential for Mycobacterium tuberculosis replication and survival during antibiotic treatment, binds to RNA polymerase (RNAP) in the absence of promoter DNA. It has been hypothesized that RbpA stimulates housekeeping gene expression by promoting assembly of the σA subunit with core RNAP. Here, using a purified in vitro transcription system of M. tuberculosis, we show that RbpA functions in a promoter-dependent manner as a companion of RNAP essential for promoter DNA unwinding and formation of the catalytically active open promoter complex (RPo). Screening for RbpA activity using a full panel of the M. tuberculosis σ subunits demonstrated that RbpA targets σA and stress-response σB, but not the alternative σ subunits from the groups 3 and 4. In contrast to σA, the σB subunit activity displayed stringent dependency upon RbpA. These results suggest that RbpA-dependent control of RPo formation provides a mechanism for tuning gene expression during the switch between different physiological states, and in the stress response.
Pavankumar, A.R., Perumal, A.S., Sankaran, K., 2012. Small RNA fragments in complex culture media cause alterations in protein profiles of three species of bacteria. BioTechniques 52, 167 MoreLess
Efforts to delineate the basis for variations in protein profiles of different membrane fractions from various bacterial pathogens led to the finding that even the same medium [e.g., Luria Bertani (LB) broth] purchased from different commercial sources generates remarkably dissimilar protein profiles despite similar growth characteristics. Given the pervasive roles small RNAs play in regulating gene expression, we inquired if these source-specific differences due to media arise from disparities in the presence of small RNAs. Indeed, LB media components from two different commercial suppliers contained varying, yet significant, amounts of 10–80 bp small RNAs. Removal of small RNA from LB using RNaseA during media preparation resulted in significant changes in bacterial protein expression profiles. Our studies underscore the fact that seemingly identical growth media can lead to dramatic alterations in protein expression patterns, highlighting the importance of utilizing media free of small RNA during bacteriological studies. Finally, these results raise the intriguing possibility that similar pools of small RNAs in the environment can influence bacterial adaptation.
Balasubramaniam, B., Perumal, A.S., Jayaraman, J., Mani, J., Ramanujam, P., 2012. Comparative analysis for the production of fatty acid alkyl esterase using whole cell biocatalyst and purified enzyme from Rhizopus oryzae on waste cooking oil (sunflower oil). Waste management 32, 1539-1547 MoreLess
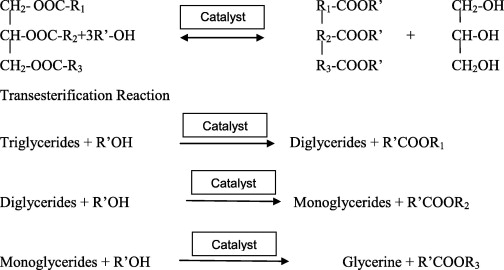 The petroleum fuel is nearing the line of extinction. Recent research and technology have provided promising outcomes to rely on biodiesel as the alternative and conventional source of fuel. The use of renewable source – vegetable oil constitutes the main stream of research. In this preliminary study, Waste Cooking Oil (WCO) was used as the substrate for biodiesel production. Lipase enzyme producing fungi Rhizopus oryzae 262 and commercially available pure lipase enzyme were used for comparative study in the production of Fatty Acid Alkyl Esters (FAAE). The whole cell (RO 262) and pure lipase enzyme (PE) were immobilized using calcium alginate beads. Calcium alginate was prepared by optimizing with different molar ratios of calcium chloride and different per cent sodium alginate. Entrapment immobilization was done for whole cell biocatalyst (WCB). PE was also immobilized by entrapment for the transesterification reaction. Seven different solvents – methanol, ethanol, n-propanol, n-butanol, iso-propanol, iso-butanol and iso-amyl alcohol were used as the acyl acceptors. The reaction parameters like temperature (30 °C), molar ratio (1:3 – oil:solvent), reaction time (24 h), and amount of enzyme (10% mass ratio to oil) were also optimized for methanol alone. The same parameters were adopted for the other acyl acceptors too. Among the different acyl acceptors – methanol, whose reaction parameters were optimized showed maximum conversion of triglycerides to FAAE-94% with PE and 84% with WCB. On the whole, PE showed better catalytic converting ability with all the acyl acceptor compared to WCB. Gas chromatography analysis (GC) was done to determine the fatty acid composition of WCO (sunflower oil) and FAAE production with different acyl acceptors.
The petroleum fuel is nearing the line of extinction. Recent research and technology have provided promising outcomes to rely on biodiesel as the alternative and conventional source of fuel. The use of renewable source – vegetable oil constitutes the main stream of research. In this preliminary study, Waste Cooking Oil (WCO) was used as the substrate for biodiesel production. Lipase enzyme producing fungi Rhizopus oryzae 262 and commercially available pure lipase enzyme were used for comparative study in the production of Fatty Acid Alkyl Esters (FAAE). The whole cell (RO 262) and pure lipase enzyme (PE) were immobilized using calcium alginate beads. Calcium alginate was prepared by optimizing with different molar ratios of calcium chloride and different per cent sodium alginate. Entrapment immobilization was done for whole cell biocatalyst (WCB). PE was also immobilized by entrapment for the transesterification reaction. Seven different solvents – methanol, ethanol, n-propanol, n-butanol, iso-propanol, iso-butanol and iso-amyl alcohol were used as the acyl acceptors. The reaction parameters like temperature (30 °C), molar ratio (1:3 – oil:solvent), reaction time (24 h), and amount of enzyme (10% mass ratio to oil) were also optimized for methanol alone. The same parameters were adopted for the other acyl acceptors too. Among the different acyl acceptors – methanol, whose reaction parameters were optimized showed maximum conversion of triglycerides to FAAE-94% with PE and 84% with WCB. On the whole, PE showed better catalytic converting ability with all the acyl acceptor compared to WCB. Gas chromatography analysis (GC) was done to determine the fatty acid composition of WCO (sunflower oil) and FAAE production with different acyl acceptors.
Bharathiraja, B., Perumal, A.S., Kalash, R.S., 2009. Enhanced production of cellulase protein on mixture of xylose, lactose, cellulose and sugarcane leaf using Trichoderma reesei 992 6a. Advanced Biotech 2009 8(07), 26-30 MoreLess
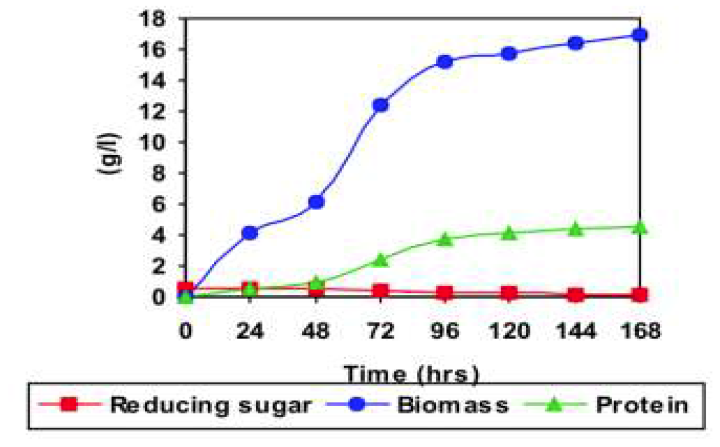 Cellulase production was studied on the mixture of xylose, lactose and cellulose then it was compared to the natural source (sugarcane leaf) by the strain Trichoderma reesei 992 6a. The experiment was conducted on single substrate i.e., xylose, lactose, cellulose and the combination of these three substrates. Then sugarcane leaf was used as another substrate. The combination of the three substrate produced highest cellulase activity and reached the maximum protein than others. When compared to the other individual substrates and natural source (sugarcane leaf). A Triauxic pattern of utilization of three carbon sources was observed as well as compared to the single and natural carbon sources. Xylose and lactose was utilized first to support growth followed by cellulose to induce the cellulase enzyme production. The mixture of xylose, lactose and cellulose used in batch enzyme production. This mixture produced the highest maximal cellulase activity of 6.4 IFPU/ml in same time than others.
Cellulase production was studied on the mixture of xylose, lactose and cellulose then it was compared to the natural source (sugarcane leaf) by the strain Trichoderma reesei 992 6a. The experiment was conducted on single substrate i.e., xylose, lactose, cellulose and the combination of these three substrates. Then sugarcane leaf was used as another substrate. The combination of the three substrate produced highest cellulase activity and reached the maximum protein than others. When compared to the other individual substrates and natural source (sugarcane leaf). A Triauxic pattern of utilization of three carbon sources was observed as well as compared to the single and natural carbon sources. Xylose and lactose was utilized first to support growth followed by cellulose to induce the cellulase enzyme production. The mixture of xylose, lactose and cellulose used in batch enzyme production. This mixture produced the highest maximal cellulase activity of 6.4 IFPU/ml in same time than others.

