Attention Everyone,
mark your calendar and watch online
Wednesday, May 25, 2016 – 4:30pm to 5:30pm
Dan V. Nicolau will present ‘Unconventional Computing’ at Stanford University.
About the talk:
Many important mathematical problems, ranging from cryptography, network routing, and protein folding, require the exploration a large number of candidate solutions. Because the time required for solving these problems grows exponentially with their size, electronic computers, which operate sequentially, cannot solve them in a reasonable timeframe. Unfortunately, the parallel-computation approaches proposed so far, e.g., DNA-, and quantum-computing, suffer from fundamental and practical drawbacks, which prevented their successful implementation. On the other hand, biological entities, from microorganisms to humans, process information in parallel, routinely, for essential tasks, such as foraging, searching for available space, competition, and cooperation. However, aside of their sheer complexity, parallel biological processes are difficult to harness for artificial parallel computation because of a fundamental difference: biological entities process analog information, e.g., concentration gradients, whereas computing devices require the processing of numbers. This subtle, but important difference between artificial and biological computation, together with the opportunity to operate biocomputation with large numbers of (small) biological agents, opens three possible avenues for development.
Biological IT. The first opportunity relies on the study of the natural procedures used by biological agents, e.g., for space search and partitioning, chemotaxis, etc., followed by the translation of these procedures in abstract mathematical algorithms. These bioinspired algorithms can be then benchmarked against standard analogues used for similar tasks, and, if appropriate, improved and implemented. Along this development avenue, which is conceptually similar to other biomimetics efforts, such as biomimetic materials, we have shown that fungi used exquisitely efficient algorithms for search for available space; and that the chemotaxis procedures used by bacteria can be used to find edges of geometrical patterns.
Biosimulation. The second opportunity relies on the capacity of using large numbers of biological agents to explore complex networks which mimic real traffic situations. This line of development has been almost entirely dedicated to the study of network optimization performed by amoeboid organisms, e.g., Physarum, placed in geometrically confined environments which also contain chemotactic ‘cues’, e.g., larger concentrations of nutrients in set coordinates. This physical simulation of traffic networks resulted in many studies assessing the optimality of real traffic networks in many countries.
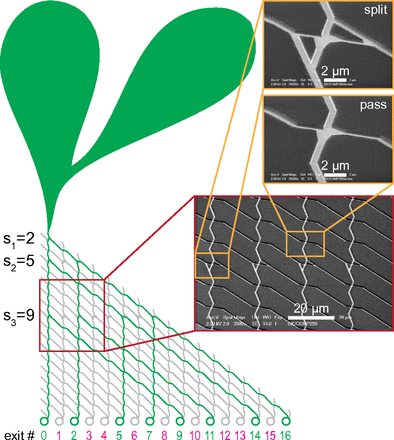
Biocomputation with biological agents in networks. Finally, the third, and arguably the most exciting development consists in the use of very large number of agents exploring purposefully-designed microfluidics networks. For instance, we reported the foundations of a parallel-computation system in which a given combinatorial problem is encoded into a graphical, modular network that is embedded in a nanofabricated planar device. Exploring the network in a parallel fashion using a large number of independent, agents, e.g., molecular motor-propelled agents, then solves the mathematical problem. This approach uses orders of magnitude less energy than conventional computers, thus addressing issues related to power consumption and heat dissipation.
The lecture will conclude with a perspective on the computation and simulation using biological entities in microfluidics structures, weighing the opportunities and challenges offered by various technological avenues.