Most recent publications
For a complete list of publications (1990 - 2016) please click here.
Hanson, K. L., Fulga, F., Dobroiu, S., Solana, G., Kašpar, O., Tokárová, V., Nicolau, D.V., Polymer surface properties control the function of heavy meromyosin in dynamic nanodevices, Biosensors and Bioelectronics (2016) MoreLess
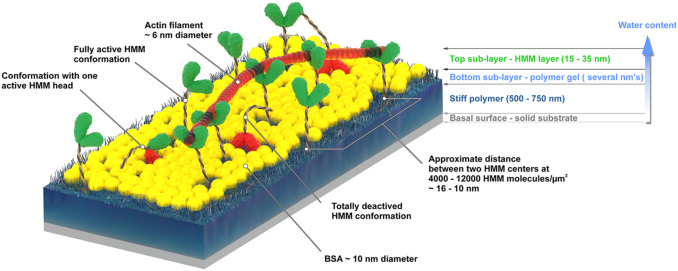
The actin-myosin system, responsible for muscle contraction, is also the force-generating element in dynamic nanodevices operating with surface-immobilized motor proteins. These devices require materials that are amenable to micro- and nano-fabrication, but also preserve the bioactivity of molecular motors. The complexity of the protein-surface systems is greatly amplified by those of the polymer-fluid interface; and of the structure and function of molecular motors, making the study of these interactions critical to the success of molecular motor-based nanodevices. We measured the density of the adsorbed motor protein (heavy meromyosin, HMM) using quartz crystal microbalance; and motor bioactivity with ATPase assay, on a set of model surfaces, i.e., nitrocellulose, polystyrene, poly(methyl methacrylate), and poly(butyl methacrylate), poly(tert-butyl methacrylate). A higher hydrophobicity of the adsorbing material translates in a higher total number of HMM molecules per unit area, but also in a lower uptake of water, and a lower ratio of active per total HMM molecules per unit area. We also measured the motility characteristics of actin filaments on the model surfaces, i.e., velocity, smoothness and deflection of movement, determined via in vitro motility assays. The filament velocities were found to be controlled by the relative number of active HMM per total motors, rather than their absolute surface density. The study allowed the formulation of the general engineering principles for the selection of polymeric materials for the manufacturing of dynamic nanodevices using protein molecular motors.
Kašpar, O., Zhang, H., Tokárová, V., Boysen, R. I., Suñé, G. R., Borrise, X., Murano, F.P., Hearn, M. T. W., Nicolau, D.V., "Confinement of water droplets on rectangular micro/nano-arrayed surfaces", Lab Chip, 16, 2487-2493 (2016) MoreLess
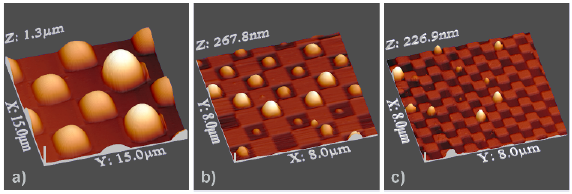
Micro-patterned surfaces with alternate hydrophilic and hydrophobic rectangular areas effectively confine water droplets down to attolitre volumes. The contact angle, volume, and geometry of the confined droplets as a function of the geometry and physico-chemical properties of the confining surfaces have been determined by phenomenological simulations, validated by atomic force microscopy measurements. The combination between experiments and simulations can be used for the purposeful design of arrays with surface-addressable hydrophobicity employed in digital microfluidics and high-throughput screening nanoarrays.
Nicolau, D. V. Jr., Lard, M., Korten, T., Van Delft, F. C. M. J. M., Persson, M., Bengtsson, E., Månsson, A., Diez, S., Linke H., Nicolau, D.V., Parallel computation with molecular-motor-propelled agents in nanofabricated networks. PNAS, 2016, doi:10.1073/pnas.1510825113 MoreLess
Asenova, E., Lin, H. Y., Fu, E., Nicolau, D. V. Jr., Nicolau, D.V. "Optimal Fungal Space Searching Algorithms", IEEE Transactions on NanoBioscience, PP, 1 (2016) MoreLess
Filipponi, L., Livingston, P., Kašpar, O., Tokárová, V., Nicolau, D.V., "Protein patterning by microcontact printing using pyramidal PDMS stamps", Biomedical Microdevices 18, 1-7 (2016) MoreLess
Nguyen, M.T., Chaffee, A.L., Boysen, R.I., Nicolau, D.V., Hearn, M.T.W., A versatile modelling approach to determine the hydrophobicity of peptides at the atomic level. Molecular Simulation, 2016. 42(4): p. 257-269 MoreLess
Van Zalinge, H., Ramsey, L.C., Aveyard, J., Persson, M., Mansson, A., Nicolau, D.V., Surface-Controlled Properties of Myosin Studied by Electric Field Modulation. Langmuir, 2015. 31(30): p. 8354-8361 MoreLess
Hajne, J., Hanson, K.L., van Zalinge, H., Nicolau, D.V., Motility of actin filaments on micro-contact printed myosin patterns. IEEE Transactions on Nanobioscience, 2015. 14(3): p. 313-322 MoreLess
Ramsey, L., Schroeder, V., van Zalinge, H., Berndt, M., Korten, T., Diez, S., Nicolau, D.V., Control and gating of kinesin-microtubule motility on electrically heated thermo-chips. Biomedical Microdevices, 2014. 16(3): p. 459-463 MoreLess
Nicolau, D.V. Jr., Paszek, E., Fulga, F., Nicolau, D.V., Mapping hydrophobicity on the protein molecular surface at atom-level resolution. PLoS ONE, 2014. 9(12) MoreLess
Wilson, R., Cossins, A., Nicolau, D. V., Missailidis, S., The selection of DNA aptamers for two different epitopes of thrombin was not due to different partitioning methods. Nucleic Acid Therapeutics, 2013. 23(1): p. 88-92 MoreLess
Nicolau, D. V., Paszek, E., Fulga, F., Nicolau Jr, D. V., Protein Molecular Surface Mapped at Different Geometrical Resolutions. PLoS ONE, 2013. 8(3) MoreLess
Bartolini, M., Naldi, M., Nicolau, D. V., Van Delft, F. C. M. J. M., Van Zijl, J., Snijder, J., Van Den Heuvel, E. F. C., Naburgh, E. P., Calonghi, N., Andrisano, V., Fluorescence biosensing micropatterned surfaces based on immobilized human acetylcholinesterase. Analytical and Bioanalytical Chemistry, 2013. 405(2-3): p. 795-804 MoreLess
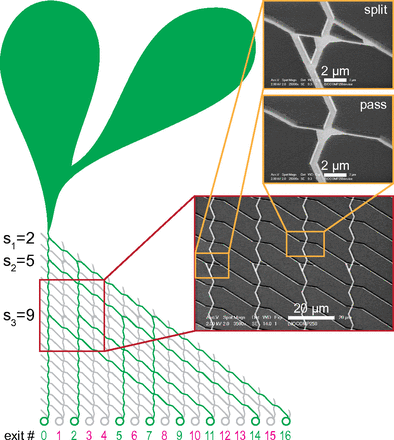 The combinatorial nature of many important mathematical problems, including nondeterministic-polynomial-time (NP)-complete problems, places a severe limitation on the problem size that can be solved with conventional, sequentially operating electronic computers. There have been significant efforts in conceiving parallel-computation approaches in the past, for example: DNA computation, quantum computation, and microfluidics-based computation. However, these approaches have not proven, so far, to be scalable and practical from a fabrication and operational perspective. Here, we report the foundations of an alternative parallel-computation system in which a given combinatorial problem is encoded into a graphical, modular network that is embedded in a nanofabricated planar device. Exploring the network in a parallel fashion using a large number of independent, molecular motor-propelled agents then solves the mathematical problem. This approach uses orders of magnitude less energy than conventional computers, thus addressing issues related to power consumption and heat dissipation. We provide a proof-of-concept demonstration of such a device by solving, in a parallel fashion, the small instance {2, 5, 9} of the subset sum problem, which is a benchmark NP-complete problem. Finally, we discuss the technical advances necessary to make our system scalable with presently available technology.
The combinatorial nature of many important mathematical problems, including nondeterministic-polynomial-time (NP)-complete problems, places a severe limitation on the problem size that can be solved with conventional, sequentially operating electronic computers. There have been significant efforts in conceiving parallel-computation approaches in the past, for example: DNA computation, quantum computation, and microfluidics-based computation. However, these approaches have not proven, so far, to be scalable and practical from a fabrication and operational perspective. Here, we report the foundations of an alternative parallel-computation system in which a given combinatorial problem is encoded into a graphical, modular network that is embedded in a nanofabricated planar device. Exploring the network in a parallel fashion using a large number of independent, molecular motor-propelled agents then solves the mathematical problem. This approach uses orders of magnitude less energy than conventional computers, thus addressing issues related to power consumption and heat dissipation. We provide a proof-of-concept demonstration of such a device by solving, in a parallel fashion, the small instance {2, 5, 9} of the subset sum problem, which is a benchmark NP-complete problem. Finally, we discuss the technical advances necessary to make our system scalable with presently available technology.
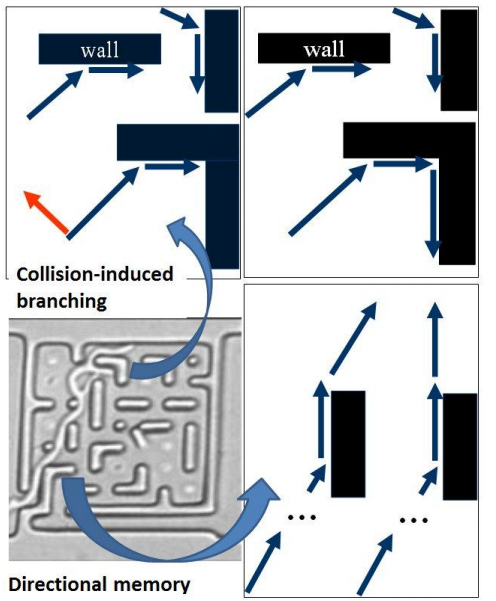 Previous experiments have shown that fungi use an efficient natural algorithm for searching the space available for their growth in micro-confined networks, e.g., mazes. This natural ‘master’ algorithm, which comprises two ‘slave’ sub-algorithms, i.e., collision-induced branching and directional memory, has been shown to be more efficient than alternatives, with one, or the other, or both sub-algorithms turned off. In contrast, the present contribution compares the performance of the fungal natural algorithm against several standard artificial homologues. It was found that the space-searching fungal algorithm consistently outperforms uninformed algorithms, such as Depth-First-Search (DFS). Furthermore, while the natural algorithm is inferior to informed ones, such as A*, this under-performance does not importantly increase with the increase of the size of the maze. These findings suggest that a systematic effort of harvesting the natural space searching algorithms used by microorganisms is warranted and possibly overdue. These natural algorithms, if efficient, can be reverse-engineered for graph and tree search strategies.
Previous experiments have shown that fungi use an efficient natural algorithm for searching the space available for their growth in micro-confined networks, e.g., mazes. This natural ‘master’ algorithm, which comprises two ‘slave’ sub-algorithms, i.e., collision-induced branching and directional memory, has been shown to be more efficient than alternatives, with one, or the other, or both sub-algorithms turned off. In contrast, the present contribution compares the performance of the fungal natural algorithm against several standard artificial homologues. It was found that the space-searching fungal algorithm consistently outperforms uninformed algorithms, such as Depth-First-Search (DFS). Furthermore, while the natural algorithm is inferior to informed ones, such as A*, this under-performance does not importantly increase with the increase of the size of the maze. These findings suggest that a systematic effort of harvesting the natural space searching algorithms used by microorganisms is warranted and possibly overdue. These natural algorithms, if efficient, can be reverse-engineered for graph and tree search strategies.
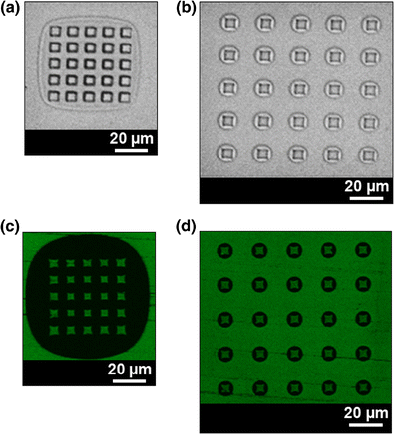 Micro-contact printing, μCP, is a well-established soft-lithography technique for printing biomolecules. μCP uses stamps made of Poly(dimethylsiloxane), PDMS, made by replicating a microstructured silicon master fabricated by semiconductor manufacturing processes. One of the problems of the μCP is the difficult control of the printing process, which, because of the high compressibility of PDMS, is very sensitive to minute changes in the applied pressure. This over-sensitive response leads to frequent and/or uncontrollable collapse of the stamps with high aspect ratios, thus decreasing the printing accuracy and reproducibility. Here we present a straightforward methodology of designing and fabricating PDMS structures with an architecture which uses the collapse of the stamp to reduce, rather than enlarge the variability of the printing. The PDMS stamp, organized as an array of pyramidal micro-posts, whose ceiling collapses when pressed on a flat surface, replicates the structure of the silicon master fabricated by anisotropic wet etching. Upon application of pressure, depending on the size of, and the pitch between, the PDMS pyramids, an air gap is formed surrounding either the entire array, or individual posts. The printing technology, which also exhibits a remarkably low background noise for fluorescence detection, may find applications when the clear demarcation of the shapes of protein patterns and the distance between them are critical, such as microarrays and studies of cell patterning.
Micro-contact printing, μCP, is a well-established soft-lithography technique for printing biomolecules. μCP uses stamps made of Poly(dimethylsiloxane), PDMS, made by replicating a microstructured silicon master fabricated by semiconductor manufacturing processes. One of the problems of the μCP is the difficult control of the printing process, which, because of the high compressibility of PDMS, is very sensitive to minute changes in the applied pressure. This over-sensitive response leads to frequent and/or uncontrollable collapse of the stamps with high aspect ratios, thus decreasing the printing accuracy and reproducibility. Here we present a straightforward methodology of designing and fabricating PDMS structures with an architecture which uses the collapse of the stamp to reduce, rather than enlarge the variability of the printing. The PDMS stamp, organized as an array of pyramidal micro-posts, whose ceiling collapses when pressed on a flat surface, replicates the structure of the silicon master fabricated by anisotropic wet etching. Upon application of pressure, depending on the size of, and the pitch between, the PDMS pyramids, an air gap is formed surrounding either the entire array, or individual posts. The printing technology, which also exhibits a remarkably low background noise for fluorescence detection, may find applications when the clear demarcation of the shapes of protein patterns and the distance between them are critical, such as microarrays and studies of cell patterning.
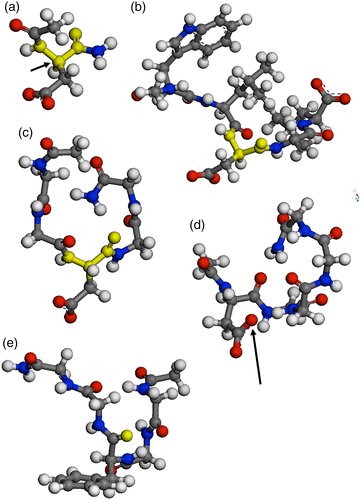 This study describes a versatile computational method to determine the hydrophobicity of small peptides at the atomic level. Free energies of transfer for individual atoms in peptide structures were derived, utilising two specifically defined parameters: (i) the water-excluding distance to define the dynamic interface between a peptide solute and its surrounding solvent and (ii) the corresponding hydrophobicity index as a relative measure for water occlusion/repulsion. The method was tested on a range of small peptide models (Ac-X-NH2, G-X-G, Ac-WL-X-LL and Ac-GG-X-GG-NH2) and several derivatives of these structures, whereby X was any of the 20 most common amino acids that naturally occur in polypeptides or proteins. The advantage of this new method lies in its versatility, ease to implement and capability to provide information on the hydrophobicity characteristics at the atomic level. The approach also encapsulates the impact of factors that influence these properties, but which have hitherto been difficult to accurately quantify, e.g. steric hindrance or proximity effects due to nearby polarised atoms. The method is not conditional on the knowledge of hydrophobicity parameters from the literature and does not require a sophisticated computer software/hardware to enable the atomic solvent-accessible surface areas or other hydrophobicity parameters to be de novo obtained.
This study describes a versatile computational method to determine the hydrophobicity of small peptides at the atomic level. Free energies of transfer for individual atoms in peptide structures were derived, utilising two specifically defined parameters: (i) the water-excluding distance to define the dynamic interface between a peptide solute and its surrounding solvent and (ii) the corresponding hydrophobicity index as a relative measure for water occlusion/repulsion. The method was tested on a range of small peptide models (Ac-X-NH2, G-X-G, Ac-WL-X-LL and Ac-GG-X-GG-NH2) and several derivatives of these structures, whereby X was any of the 20 most common amino acids that naturally occur in polypeptides or proteins. The advantage of this new method lies in its versatility, ease to implement and capability to provide information on the hydrophobicity characteristics at the atomic level. The approach also encapsulates the impact of factors that influence these properties, but which have hitherto been difficult to accurately quantify, e.g. steric hindrance or proximity effects due to nearby polarised atoms. The method is not conditional on the knowledge of hydrophobicity parameters from the literature and does not require a sophisticated computer software/hardware to enable the atomic solvent-accessible surface areas or other hydrophobicity parameters to be de novo obtained.
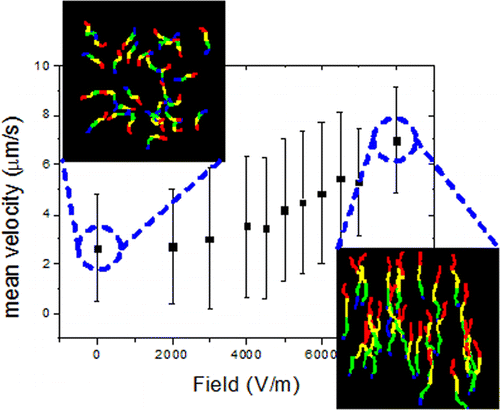 The efficiency of dynamic nanodevices using surface-immobilized protein molecular motors, which have been proposed for diagnostics, drug discovery, and biocomputation, critically depends on the ability to precisely control the motion of motor-propelled, individual cytoskeletal filaments transporting cargo to designated locations. The efficiency of these devices also critically depends on the proper function of the propelling motors, which is controlled by their interaction with the surfaces they are immobilized on. Here we use a microfluidic device to study how the motion of the motile elements, i.e., actin filaments propelled by heavy mero-myosin (HMM) motor fragments immobilized on various surfaces, is altered by the application of electrical loads generated by an external electric field with strengths ranging from 0 to 8 kVm–1. Because the motility is intimately linked to the function of surface-immobilized motors, the study also showed how the adsorption properties of HMM on various surfaces, such as nitrocellulose (NC), trimethylclorosilane (TMCS), poly(methyl methacrylate) (PMMA), poly(tert-butyl methacrylate) (PtBMA), and poly(butyl methacrylate) (PBMA), can be characterized using an external field. It was found that at an electric field of 5 kVm–1 the force exerted on the filaments is sufficient to overcome the frictionlike resistive force of the inactive motors. It was also found that the effect of assisting electric fields on the relative increase in the sliding velocity was markedly higher for the TMCS-derivatized surface than for all other polymer-based surfaces. An explanation of this behavior, based on the molecular rigidity of the TMCS-on-glass surfaces as opposed to the flexibility of the polymer-based ones, is considered. To this end, the proposed microfluidic device could be used to select appropriate surfaces for future lab-on-a-chip applications as illustrated here for the almost ideal TMCS surface. Furthermore, the proposed methodology can be used to gain fundamental insights into the functioning of protein molecular motors, such as the force exerted by the motors under different operational conditions.
The efficiency of dynamic nanodevices using surface-immobilized protein molecular motors, which have been proposed for diagnostics, drug discovery, and biocomputation, critically depends on the ability to precisely control the motion of motor-propelled, individual cytoskeletal filaments transporting cargo to designated locations. The efficiency of these devices also critically depends on the proper function of the propelling motors, which is controlled by their interaction with the surfaces they are immobilized on. Here we use a microfluidic device to study how the motion of the motile elements, i.e., actin filaments propelled by heavy mero-myosin (HMM) motor fragments immobilized on various surfaces, is altered by the application of electrical loads generated by an external electric field with strengths ranging from 0 to 8 kVm–1. Because the motility is intimately linked to the function of surface-immobilized motors, the study also showed how the adsorption properties of HMM on various surfaces, such as nitrocellulose (NC), trimethylclorosilane (TMCS), poly(methyl methacrylate) (PMMA), poly(tert-butyl methacrylate) (PtBMA), and poly(butyl methacrylate) (PBMA), can be characterized using an external field. It was found that at an electric field of 5 kVm–1 the force exerted on the filaments is sufficient to overcome the frictionlike resistive force of the inactive motors. It was also found that the effect of assisting electric fields on the relative increase in the sliding velocity was markedly higher for the TMCS-derivatized surface than for all other polymer-based surfaces. An explanation of this behavior, based on the molecular rigidity of the TMCS-on-glass surfaces as opposed to the flexibility of the polymer-based ones, is considered. To this end, the proposed microfluidic device could be used to select appropriate surfaces for future lab-on-a-chip applications as illustrated here for the almost ideal TMCS surface. Furthermore, the proposed methodology can be used to gain fundamental insights into the functioning of protein molecular motors, such as the force exerted by the motors under different operational conditions.
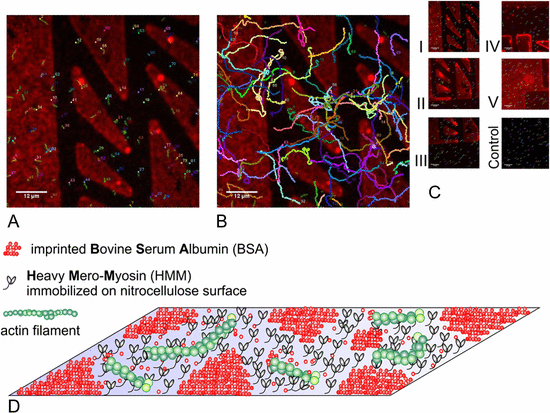 Protein molecular motors, which convert, directly and efficiently, chemical energy into motion, are excellent candidates for integration in hybrid dynamic nanodevices. To integrate and use the full potential of molecular motors in these devices, their design requires a quantitative and precise prediction of the fundamental mechanical and physicochemical features of cytoskeletal proteins operating in artificial environments. In that regard, the behavior of protein molecular motors constructs in/on nano-confined spaces or nanostructured surfaces that aim to control their motility is of critical interest. Here, we used a standard gliding motility assay to study the actin filaments sliding on a surface comprising heavy mero myosin (HMM) micro- and nano-patterns. To print HMM, we used negative tone, micro contact printing of a blocking protein (bovine serum albumin, BSA) on a nitrocellulose surface, followed by specific adsorption of HMM on BSA-free surfaces. While the large BSA-free patterns allowed for selective confinement of actin filaments motility, the BSA-stamped areas displayed intricate nano-sized HMM patterns, which enabled a deeper analysis of the nano-mechanics of actomyosin motility in confined spaces.
Protein molecular motors, which convert, directly and efficiently, chemical energy into motion, are excellent candidates for integration in hybrid dynamic nanodevices. To integrate and use the full potential of molecular motors in these devices, their design requires a quantitative and precise prediction of the fundamental mechanical and physicochemical features of cytoskeletal proteins operating in artificial environments. In that regard, the behavior of protein molecular motors constructs in/on nano-confined spaces or nanostructured surfaces that aim to control their motility is of critical interest. Here, we used a standard gliding motility assay to study the actin filaments sliding on a surface comprising heavy mero myosin (HMM) micro- and nano-patterns. To print HMM, we used negative tone, micro contact printing of a blocking protein (bovine serum albumin, BSA) on a nitrocellulose surface, followed by specific adsorption of HMM on BSA-free surfaces. While the large BSA-free patterns allowed for selective confinement of actin filaments motility, the BSA-stamped areas displayed intricate nano-sized HMM patterns, which enabled a deeper analysis of the nano-mechanics of actomyosin motility in confined spaces.
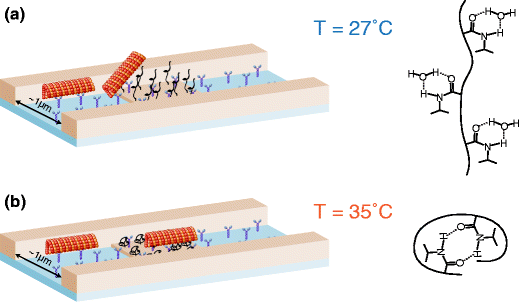 First lab-on-chip devices based on active transport by biomolecular motors have been demonstrated for basic detection and sorting applications. However, to fully employ the advantages of such hybrid nanotechnology, versatile spatial and temporal control mechanisms are required. Using a thermo-responsive polymer, we demonstrated a temperature controlled gate that either allows or disallows the passing of microtubules through a topographically defined channel. The gate is addressed by a narrow gold wire, which acts as a local heating element. It is shown that the electrical current flowing through a narrow gold channel can control the local temperature and as a result the conformation of the polymer. This is the first demonstration of a spatially addressable gate for microtubule motility which is a key element of nanodevices based on biomolecular motors.
First lab-on-chip devices based on active transport by biomolecular motors have been demonstrated for basic detection and sorting applications. However, to fully employ the advantages of such hybrid nanotechnology, versatile spatial and temporal control mechanisms are required. Using a thermo-responsive polymer, we demonstrated a temperature controlled gate that either allows or disallows the passing of microtubules through a topographically defined channel. The gate is addressed by a narrow gold wire, which acts as a local heating element. It is shown that the electrical current flowing through a narrow gold channel can control the local temperature and as a result the conformation of the polymer. This is the first demonstration of a spatially addressable gate for microtubule motility which is a key element of nanodevices based on biomolecular motors.
A precise representation of the spatial distribution of hydrophobicity, hydrophilicity and charges on the molecular surface of proteins is critical for the understanding of the interaction with small molecules and larger systems. The representation of hydrophobicity is rarely done at atom-level, as this property is generally assigned to residues. A new methodology for the derivation of atomic hydrophobicity from any amino acid-based hydrophobicity scale was used to derive 8 sets of atomic hydrophobicities, one of which was used to generate the molecular surfaces for 35 proteins with convex structures, 5 of which, i.e., lysozyme, ribonuclease, hemoglobin, albumin and IgG, have been analyzed in more detail. Sets of the molecular surfaces of the model proteins have been constructed using spherical probes with increasingly large radii, from 1.4 to 20 Å, followed by the quantification of (i) the surface hydrophobicity; (ii) their respective molecular surface areas, i.e., total, hydrophilic and hydrophobic area; and (iii) their relative densities, i.e., divided by the total molecular area; or specific densities, i.e., divided by property-specific area. Compared with the amino acid-based formalism, the atom-level description reveals molecular surfaces which (i) present an approximately two times more hydrophilic areas; with (ii) less extended, but between 2 to 5 times more intense hydrophilic patches; and (iii) 3 to 20 times more extended hydrophobic areas. The hydrophobic areas are also approximately 2 times more hydrophobicity-intense. This, more pronounced “leopard skin”-like, design of the protein molecular surface has been confirmed by comparing the results for a restricted set of homologous proteins, i.e., hemoglobins diverging by only one residue (Trp37). These results suggest that the representation of hydrophobicity on the protein molecular surfaces at atom-level resolution, coupled with the probing of the molecular surface at different geometric resolutions, can capture processes that are otherwise obscured to the amino acid-based formalism.
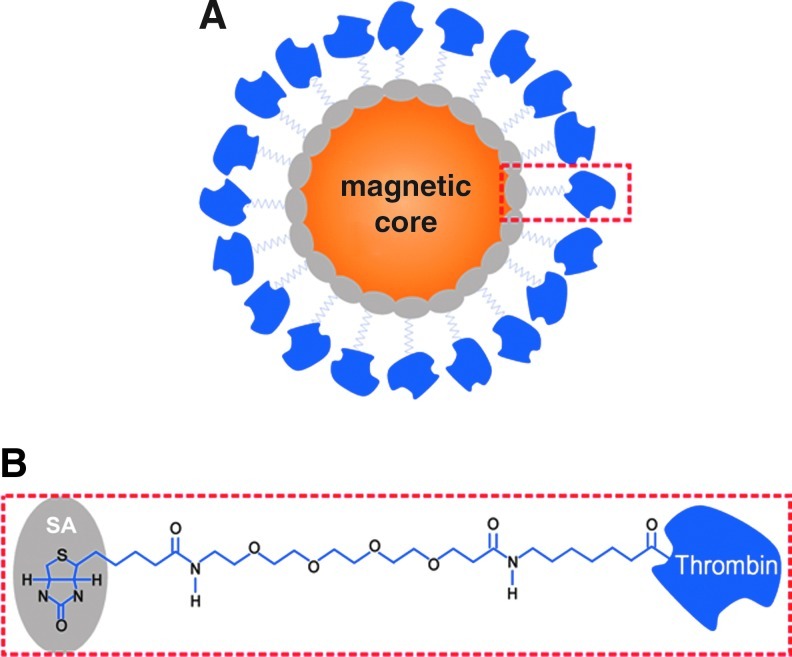 Nearly all aptamers identified so far for any given target molecule have been specific for the same binding site (epitope). The most notable exception to the 1 aptamer per target molecule rule is the pair of DNA aptamers that bind to different epitopes of thrombin. This communication refutes the suggestion that these aptamers exist because different partitioning methods were used when they were selected. The possibility that selection of these aptamers was biased by conflicting secondary structures was also investigated and found not to contribute. The preparation of protein-coated magnetic beads for systematic evolution of ligands by exponential enrichment (SELEX) and the different specificities of the thrombin aptamers for the α and β forms of thrombin are also reported.
Nearly all aptamers identified so far for any given target molecule have been specific for the same binding site (epitope). The most notable exception to the 1 aptamer per target molecule rule is the pair of DNA aptamers that bind to different epitopes of thrombin. This communication refutes the suggestion that these aptamers exist because different partitioning methods were used when they were selected. The possibility that selection of these aptamers was biased by conflicting secondary structures was also investigated and found not to contribute. The preparation of protein-coated magnetic beads for systematic evolution of ligands by exponential enrichment (SELEX) and the different specificities of the thrombin aptamers for the α and β forms of thrombin are also reported.
Many areas of biochemistry and molecular biology, both fundamental and applications-orientated, require an accurate construction, representation and understanding of the protein molecular surface and its interaction with other, usually small, molecules. There are however many situations when the protein molecular surface gets in physical contact with larger objects, either biological, such as membranes, or artificial, such as nanoparticles. The contribution presents a methodology for describing and quantifying the molecular properties of proteins, by geometrical and physico-chemical mapping of the molecular surfaces, with several analytical relationships being proposed for molecular surface properties. The relevance of the molecular surface-derived properties has been demonstrated through the calculation of the statistical strength of the prediction of protein adsorption. It is expected that the extension of this methodology to other phenomena involving proteins near solid surfaces, in particular the protein interaction with nanoparticles, will result in important benefits in the understanding and design of protein-specific solid surfaces.
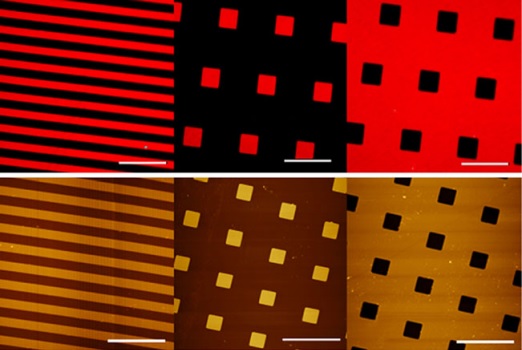 Human acetylcholinesterase (AChE) is a widely studied target enzyme in drug discovery for Alzheimer's disease (AD). In this paper we report evaluation of the optimum structure and chemistry of the supporting material for a new AChE-based fluorescence sensing surface. To achieve this objective, multilayered silicon wafers with spatially controlled geometry and chemical diversity were fabricated. Specifically, silicon wafers with silicon oxide patterns (SiO(2)/Si wafers), platinum-coated silicon wafers with SiO(2) patterns (SiO(2)/Pt/Ti/Si wafers), and Pt-coated wafers coated with different thicknesses of TiO(2) and SiO(2) (SiO(2)/TiO(2)/Pt/Ti/Si wafers) were labelled with the fluorescent conjugation agent HiLyte Fluor 555. Selection of a suitable material and the optimum pattern thickness required to maximize the fluorescence signal and maintain chemical stability was performed by confocal laser-scanning microscopy (CLSM). Results showed that the highest signal-to-background ratio was always obtained on wafers with 100 nm thick SiO(2) features. Hence, these wafers were selected for covalent binding of human AChE. Batch-wise kinetic studies revealed that enzyme activity was retained after immobilization. Combined use of atomic-force microscopy and CLSM revealed that AChE was homogeneously and selectively distributed on the SiO(2) microstructures at a suitable distance from the reflective surface. In the optimum design, efficient fluorescence emission was obtained from the AChE-based biosensing surface after labelling with propidium, a selective fluorescent probe of the peripheral binding site of AChE.
Human acetylcholinesterase (AChE) is a widely studied target enzyme in drug discovery for Alzheimer's disease (AD). In this paper we report evaluation of the optimum structure and chemistry of the supporting material for a new AChE-based fluorescence sensing surface. To achieve this objective, multilayered silicon wafers with spatially controlled geometry and chemical diversity were fabricated. Specifically, silicon wafers with silicon oxide patterns (SiO(2)/Si wafers), platinum-coated silicon wafers with SiO(2) patterns (SiO(2)/Pt/Ti/Si wafers), and Pt-coated wafers coated with different thicknesses of TiO(2) and SiO(2) (SiO(2)/TiO(2)/Pt/Ti/Si wafers) were labelled with the fluorescent conjugation agent HiLyte Fluor 555. Selection of a suitable material and the optimum pattern thickness required to maximize the fluorescence signal and maintain chemical stability was performed by confocal laser-scanning microscopy (CLSM). Results showed that the highest signal-to-background ratio was always obtained on wafers with 100 nm thick SiO(2) features. Hence, these wafers were selected for covalent binding of human AChE. Batch-wise kinetic studies revealed that enzyme activity was retained after immobilization. Combined use of atomic-force microscopy and CLSM revealed that AChE was homogeneously and selectively distributed on the SiO(2) microstructures at a suitable distance from the reflective surface. In the optimum design, efficient fluorescence emission was obtained from the AChE-based biosensing surface after labelling with propidium, a selective fluorescent probe of the peripheral binding site of AChE.
Journal publications