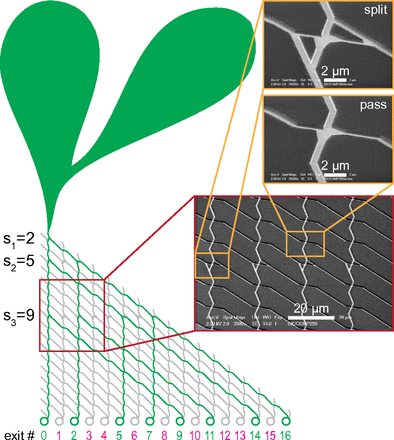
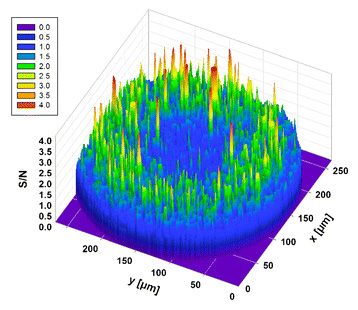
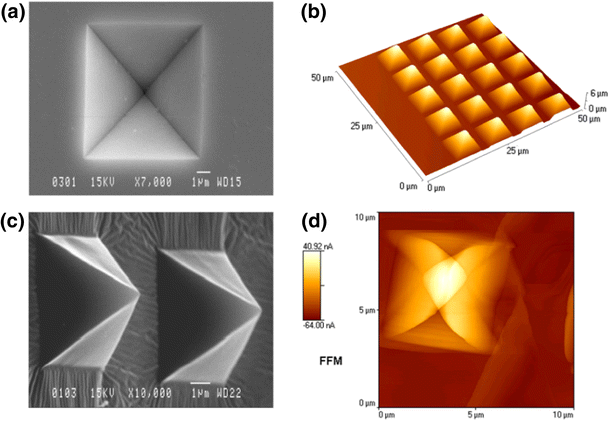
Our new paper has been just published in Biosensors and Bioelectronics!
Hanson, K. L., Fulga, F., Dobroiu, S., Solana, G., Kašpar, O., Tokárová, V., Nicolau, D. V.
Polymer surface properties control the function of heavy meromyosin in dynamic nanodevices
Abstract
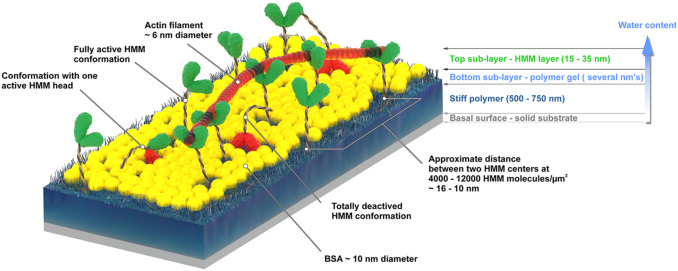 The actin-myosin system, responsible for muscle contraction, is also the force-generating element in dynamic nanodevices operating with surface-immobilized motor proteins. These devices require materials that are amenable to micro- and nano-fabrication, but also preserve the bioactivity of molecular motors. The complexity of the protein-surface systems is greatly amplified by those of the polymer-fluid interface; and of the structure and function of molecular motors, making the study of these interactions critical to the success of molecular motor-based nanodevices. We measured the density of the adsorbed motor protein (heavy meromyosin, HMM) using quartz crystal microbalance; and motor bioactivity with ATPase assay, on a set of model surfaces, i.e., nitrocellulose, polystyrene, poly(methyl methacrylate), and poly(butyl methacrylate), poly(tert-butyl methacrylate). A higher hydrophobicity of the adsorbing material translates in a higher total number of HMM molecules per unit area, but also in a lower uptake of water, and a lower ratio of active per total HMM molecules per unit area. We also measured the motility characteristics of actin filaments on the model surfaces, i.e., velocity, smoothness and deflection of movement, determined via in vitro motility assays. The filament velocities were found to be controlled by the relative number of active HMM per total motors, rather than their absolute surface density. The study allowed the formulation of the general engineering principles for the selection of polymeric materials for the manufacturing of dynamic nanodevices using protein molecular motors.
The actin-myosin system, responsible for muscle contraction, is also the force-generating element in dynamic nanodevices operating with surface-immobilized motor proteins. These devices require materials that are amenable to micro- and nano-fabrication, but also preserve the bioactivity of molecular motors. The complexity of the protein-surface systems is greatly amplified by those of the polymer-fluid interface; and of the structure and function of molecular motors, making the study of these interactions critical to the success of molecular motor-based nanodevices. We measured the density of the adsorbed motor protein (heavy meromyosin, HMM) using quartz crystal microbalance; and motor bioactivity with ATPase assay, on a set of model surfaces, i.e., nitrocellulose, polystyrene, poly(methyl methacrylate), and poly(butyl methacrylate), poly(tert-butyl methacrylate). A higher hydrophobicity of the adsorbing material translates in a higher total number of HMM molecules per unit area, but also in a lower uptake of water, and a lower ratio of active per total HMM molecules per unit area. We also measured the motility characteristics of actin filaments on the model surfaces, i.e., velocity, smoothness and deflection of movement, determined via in vitro motility assays. The filament velocities were found to be controlled by the relative number of active HMM per total motors, rather than their absolute surface density. The study allowed the formulation of the general engineering principles for the selection of polymeric materials for the manufacturing of dynamic nanodevices using protein molecular motors.